library(rstac)
library(gdalcubes)
library(stars)
library(tmap)STAC Tiling
Describing geospatial assets using STAC metadata, as with the Biodiversity Data Catalog, is not merely a convenient way to improve discovery of existing resources – it can also open the door to substantial performance gains. This is most visible when analyses involve large numbers of individual spatial assets, such as with satellite imagery.
Most geospatial formats, like geotif, embed spatial metadata such as the bounding box, description of bands, spatial projection and resolution into each individual file. Even though range request methods allow us to read these metadata bytes without downloading the entire asset, making such requests over thousands or hundreds of thousands of assets that comprise modern Earth Observation (EO) data quickly becomes a bottleneck. The metadata available in the STAC catalog for all assets (i.e. tif files) allows us to programmatically identify which assets fall into a particular space-time data cube very efficiently.
Here we illustrate the use of STAC in programmatically constructing a data cube of satellite imagery assets corresponding to the desired set of space, time, and spectral bands, and excluding assets with insufficient quality flags in metadata (in this case, above a given threshold for cloud cover.) This high-level interface allows us to ignore the nitty gritty of individual satellite image files from different bands, taken at different times, and overlapping in different parts of space, and automatically presents that information to us as a coherent data cube. The approach leverages ‘lazy evaluation’ very effectively, allowing us to avoid unnecessary computation while processing data that may be far too large to fit into computer RAM. This approach works similarly in both R and python.
R
Here we compute NDVI from Sentinel2 over arbitrary data cube:
We will search the STAC catalog for all satellite images in our desired space & time cube:
## STAC Search over 400 million assets.
box <- c(xmin=-122.51006, ymin=37.70801, xmax=-122.36268, ymax=37.80668)
start_date <- "2022-06-01"
end_date <- "2022-08-01"
items <-
stac("https://earth-search.aws.element84.com/v0/") |>
stac_search(collections = "sentinel-s2-l2a-cogs",
bbox = box,
datetime = paste(start_date, end_date, sep="/"),
limit = 100) |>
post_request() We create an image collection of those features that have our desired bands, including the cloud image mask, and filtering out images with over 20% cloud covered (as denoted in their stac metadata)
col <-
stac_image_collection(items$features,
asset_names = c("B04","B08", "SCL"),
property_filter = \(x) {x[["eo:cloud_cover"]] < 20})We can now define our cube of interest. Note that this specification is completely independent of the actual data assets – the actual spatial and temporal resolution of the data could be finer or coarser than we request! A given x,y,t pixel in this abstract cube may be covered by multiple images (e.g. overlapping images, or multiple satellite fly-bys in the same week) – and the code will aggregate them by the desired aggregation method (median). Other such x,y,t pixels in the cube may have no data coverage at all – perhaps the only images covering those pixels were removed by the cloud mask. In this case, the pixels could be filled in by the resampling method (such as a spline, nearest-neighbor, or in this case, average of nearby pixels, See gdalwarp. Note that this approach means we can define both the projection and the spatio-temporal resolution of the output completely independently from the input.
cube <- cube_view(srs = "EPSG:4326",
extent = list(t0 = start_date, t1 = end_date,
left = box[1], right = box[3],
top = box[4], bottom = box[2]),
nx = 2400, ny = 2400, dt = "P1D",
aggregation = "median", resampling = "average")
S2.mask <- image_mask("SCL", values=c(3,8,9)) # mask clouds and cloud shadows
virtual_cube <- raster_cube(col, cube, mask = S2.mask)Observe that this evaluation is “lazy”, we haven’t yet downloaded a single pixel of satellite imagery. Using our virtual cube, we can reference just the bands of interest and perform arbitrary operations on the those pixels, e.g. to calculate NDVI. This entire calculation happens over network interface range requests, first computing the aggregation and resampling warps. See the gdalcubes paper for details.
ndvi <- virtual_cube |>
select_bands(c("B04", "B08")) |>
apply_pixel("(B08-B04)/(B08+B04)", "NDVI") |>
reduce_time(c("mean(NDVI)")) |>
st_as_stars()
tm_shape(ndvi) + tm_raster(palette = viridisLite::mako(20), n=20)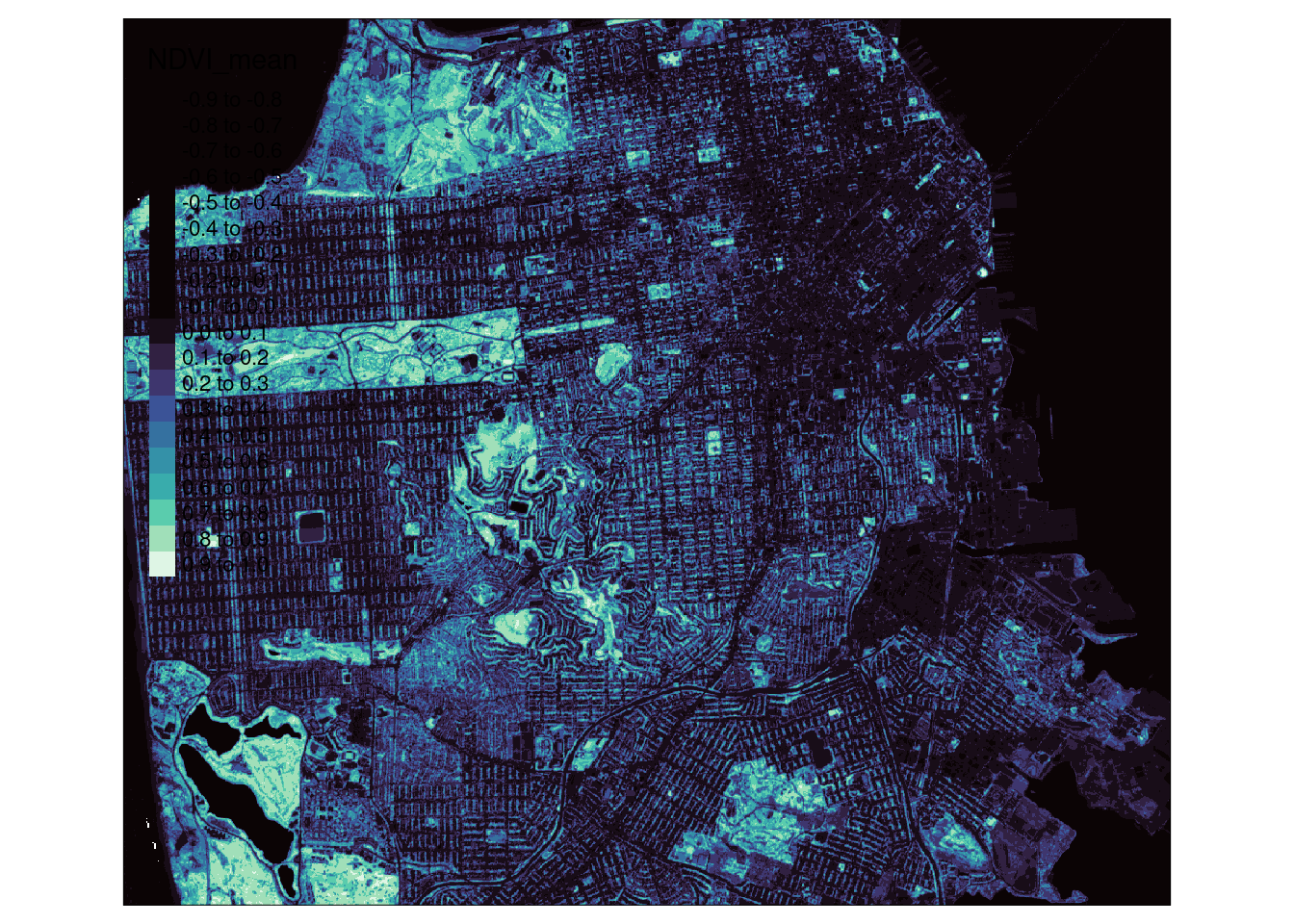
Python
pystac + stackstac provides a comparable approach to rstac + gdalcubes approach.
import pystac
import stackstac