Sys.setenv("AWS_S3_ENDPOINT"="minio.carlboettiger.info")
Sys.setenv("AWS_VIRTUAL_HOSTING"="FALSE")cloud basics
Much scientific computing focuses on the premise that data must be downloaded to a local filesystem (hard-drive) before use. This tutorial focuses on techniques for accessing data directly over a network connection to an object store (such as AWS S3, Google Storage, or Azure blobstore) instead. Object stores are not restricted to commercial cloud providers – today, many research compute centers and academic institutions host their own object stores on their own clusters, such as the NSF-funded OpenStorageNetwork, Jetstream2, using open source software like MINIO or Redhat CEPH. These open source options implement the same application programming interface as AWS S3, making them compatible with software written for this first big commercial and most widely used object store.
You do not need an actual cloud host to use these patterns – a user could install a MINIO instance on their own laptop. This can be very useful for testing purposes. Setting up MINIO instance on a personal desktop or local server can be an effective way to share data with a larger team. While this pattern is similar to relational databases such as MySQL or Postgres where data is typically accessed on a central server, it is important to note that object stores are a form of ‘static’ storage. When we make a request against a relational database asking for the sum of a slice of data, that computation is performed by the server, and only the resulting sum is sent back to our local machine. When we make such a request against an object store, the slice is extracted from the larger dataset and sent across the network, and the sum is computed on the local machine (streaming).
R
Rasters
library(sf)
library(stars)
library(rstac)
library(gdalcubes)library(dplyr)
library(spData)
india <- spData::world |> filter(name_long=="India") Single raster:
url <- "/vsis3/public-biodiversity/carbon/Irrecoverable_Carbon_2010/Irrecoverable_C_Total_2010.tif"Both stars and terra handle lazy reads well. Stars remains ‘lazy’ even after cropping, while terra evaluates at first call to crop.
r <- stars::read_stars(url)
r_cropped <- r |> st_crop(india)
plot(r_cropped)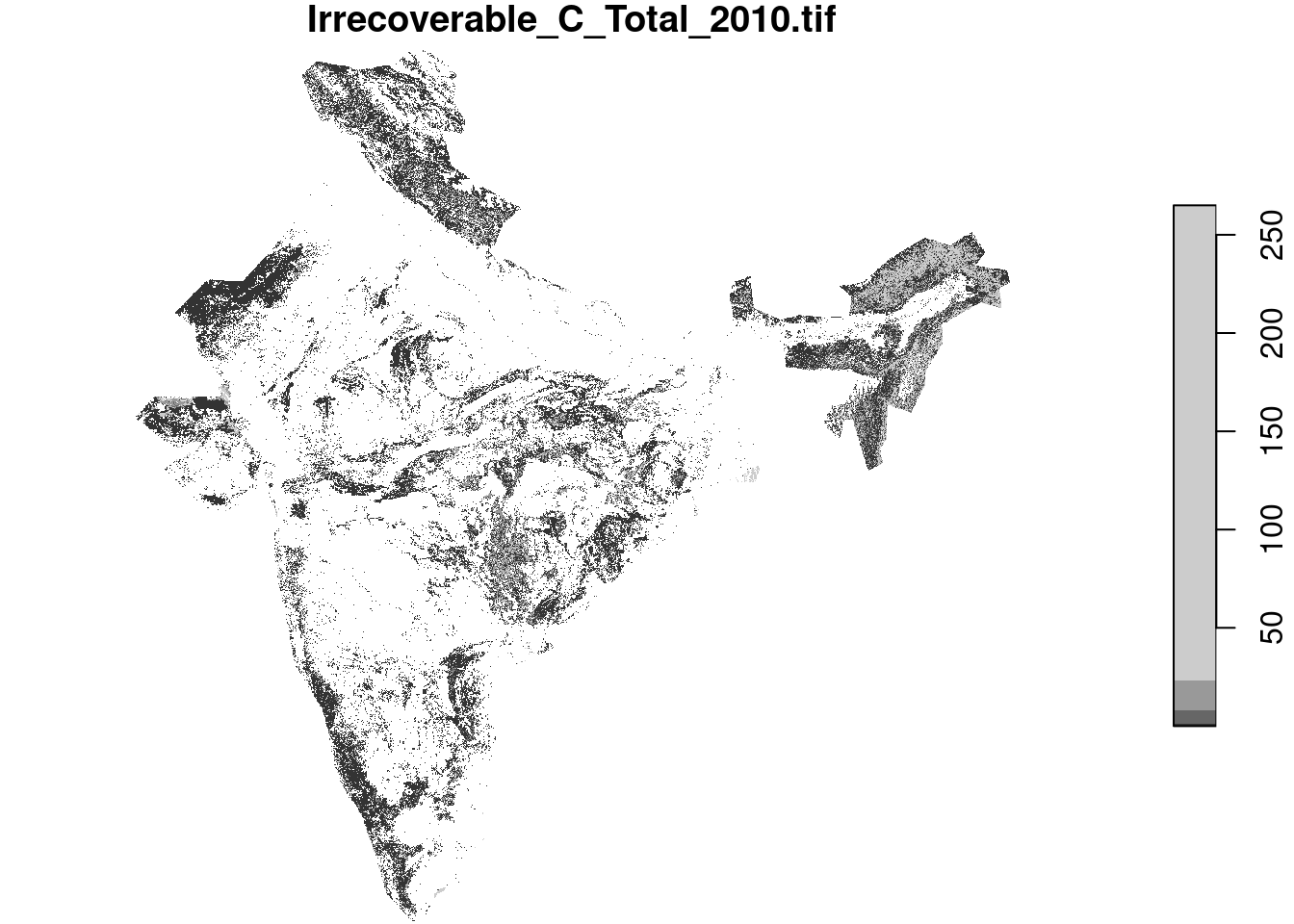
terra
library(terra)
r <- terra::rast(url)
r_cropped <- r |> terra::crop(vect(india))
plot(r_cropped)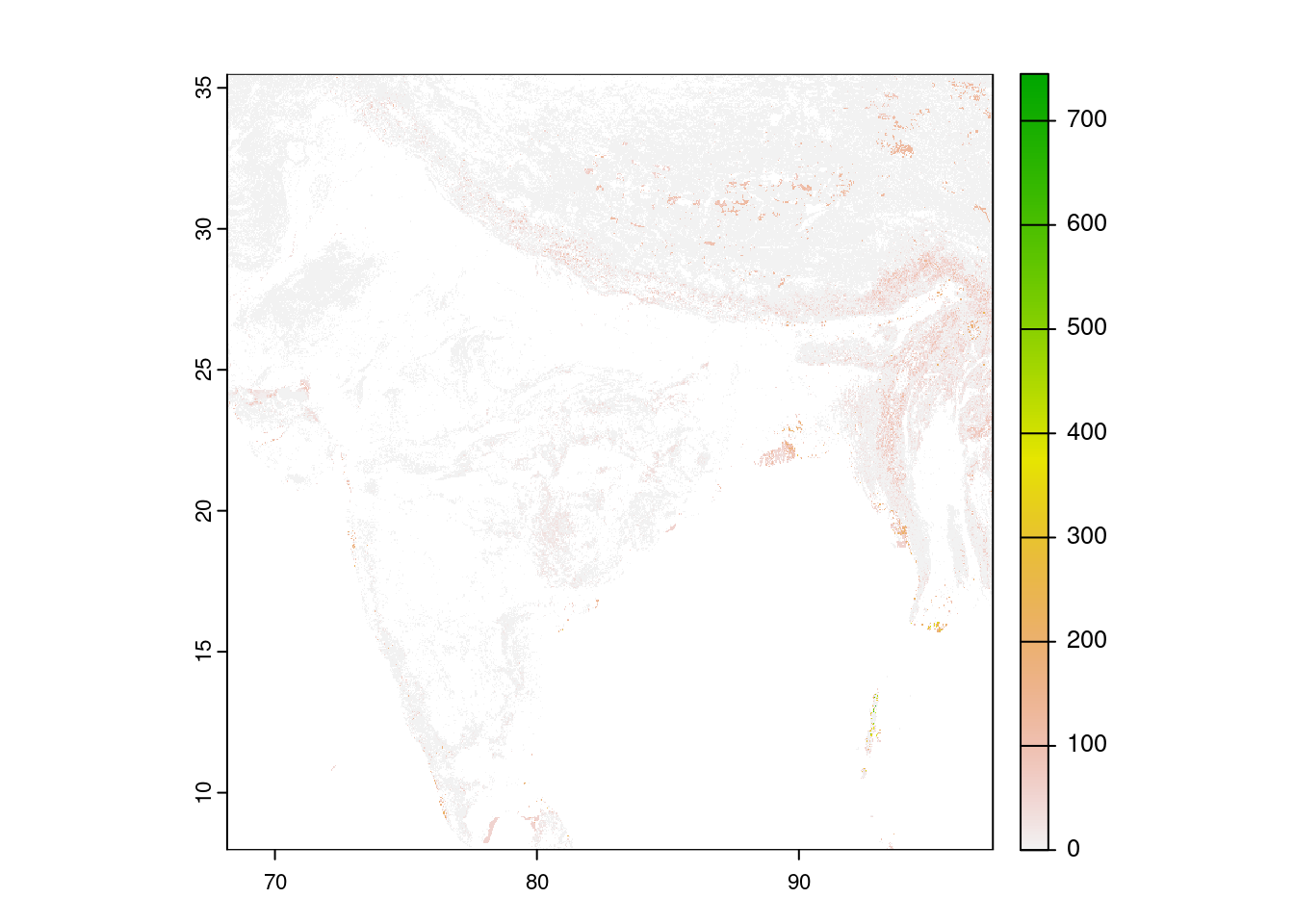
gdalcubes
gdalcubes is most powerful in working with large numbers of individual raster assets that are to be combined into a space-time “data cube”. However, it works very nicely even with a single or small collection of rasters as a high-level interface for lazy reads, including cropping, coordinate transforms, and upscaling or downscaling spatial sampling. This approach is also substantially faster than the alternative methods for remote subsetting and lazy eval analysis. See the entry on stac tiling for more details.
box <- st_bbox(india)
view <- cube_view(srs = "EPSG:4326",
extent = list(t0 = "2010-01-01", t1 = "2010-01-01",
left = box[1], right = box[3],
top = box[4], bottom = box[2]),
nx = 400, ny=400, dt = "P1Y")
img <- gdalcubes::create_image_collection(url, date_time = "2010-01-01")
raster_cube(img, view) |> plot(col = viridisLite::viridis)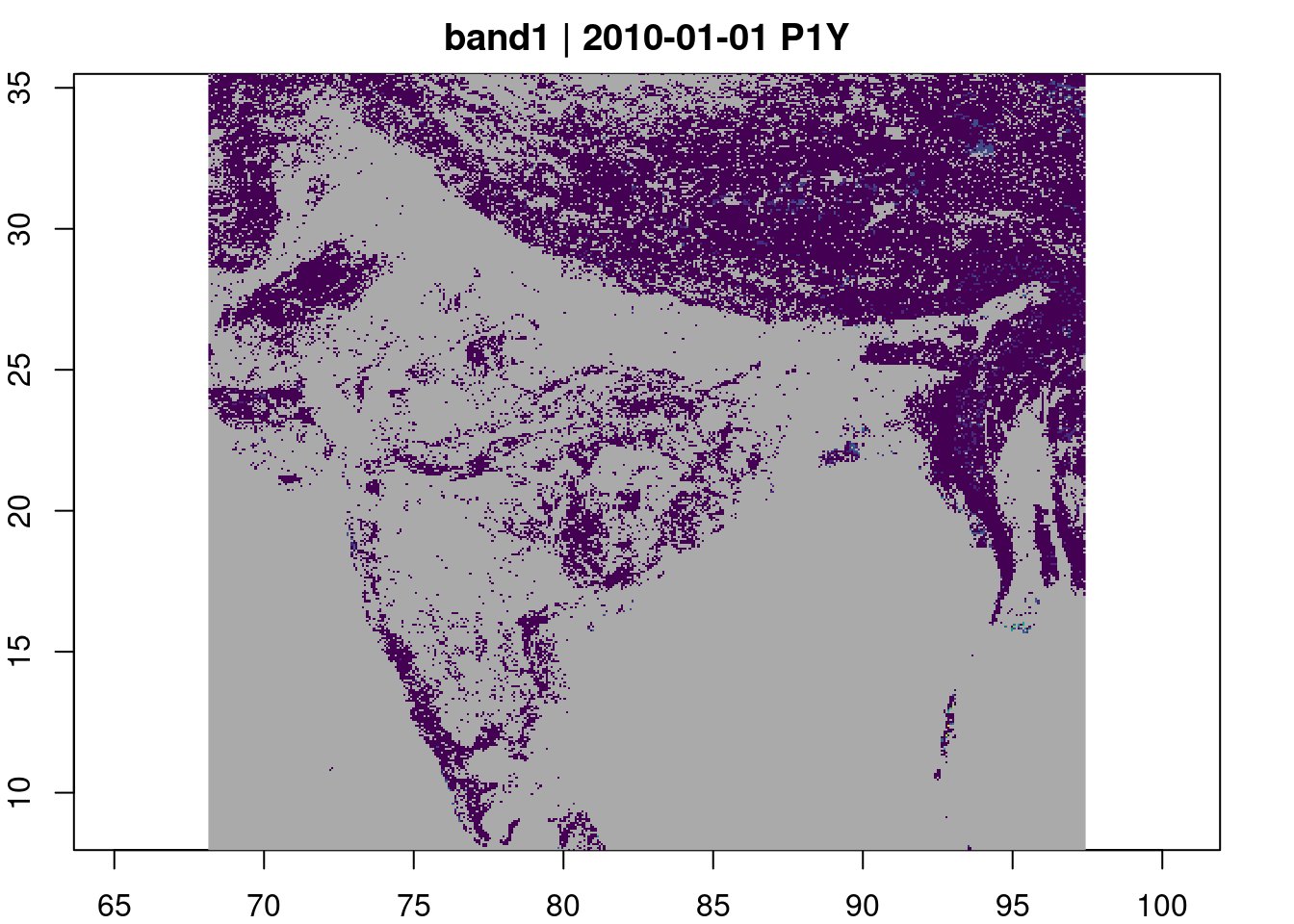
Like terra and stars, gdalcubes includes a variety of functions for pixel-based operations across different bands, often with better performance through optimized lazy-evaluation. Otherwise, use stars::st_as_stars() to coerce a raster_cube object to a stars object (e.g. for tmap and other plotting.)
Write operations
In many spatial workflows it desirable to write out intermediate or final products to files that can be rendered for visualization or consumed by other software or workflows. Just as we can read directly from our S3 cloud storage systems, we can also write our outputs back to them. See the entry on Publishing Data for details.
Vectors
Sub-setting spatial vector objects works a little differently. In both terra and sf, it is best to specify sub-setting when first opening the vector files (recent versions of terra let us delay this a bit).
wpa_url <- "/vsis3/biodiversity/World-Protected-Areas-May2023.gdb"terra
wpas <- vect(wpa_url, filter = vect(india))We can remotely filter polygon data by other columns as well by using postgis query syntax:
names <- vect(wpa_url, query = "SELECT DISTINCT NAME FROM WDPA_WDOECM_poly_May2023_all")
head(names$NAME)[1] "Hollis Road" "Wairakau" "Waihou Forest" "Wairere Falls"
[5] "Upland Rd" "Waitioka Stream"sf
sf syntax is functionally equivalent, but takes the filter as wkt text.
wpas2 <- st_read(wpa_url, wkt = st_as_text(st_geometry(india)))Plots combining vector and raster layers with tmap
library(tmap)
r <- raster_cube(img, view) |> st_as_stars()
tm_shape(r) + tm_raster(legend.show = FALSE) +
tm_shape(st_as_sf(wpas)) + tm_polygons(alpha=0.5)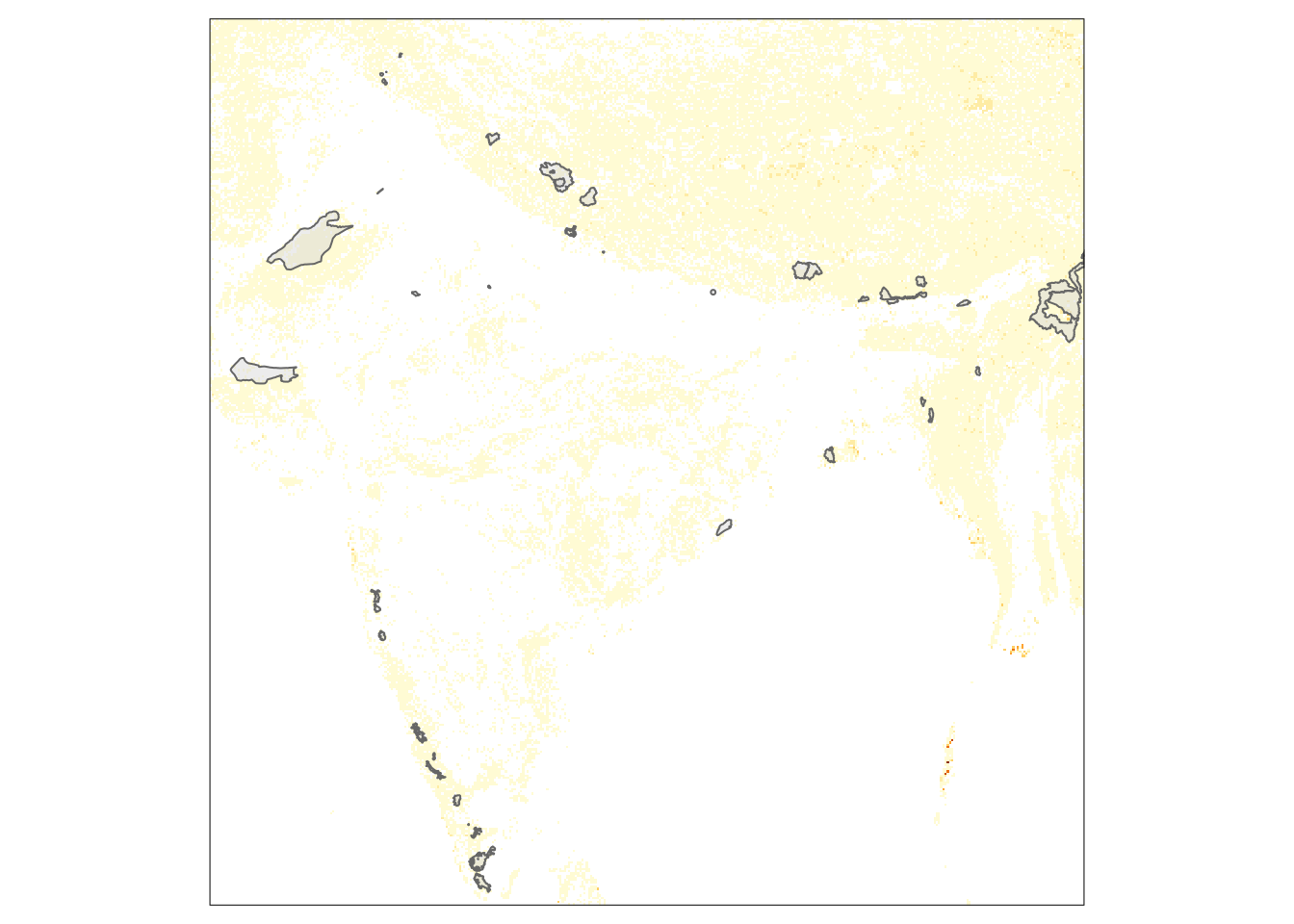
Python
(Coming soon)